
In real estate, spatial data is the name of the game. Countless programs
in other domains utilize the power of this data, which is becoming more
prevalent by the day.
In this post I will go over a few simple, but powerful tools to get you
started using using geographic information in R.
##First, some libraries##
#install.packages('GISTools', dependencies = T)
library(GISTools)
GISTools provides an easy-to-use method for creating shading schemes
and choropleth maps. Some of you may have heard of the sp package,
which adds numerous spatial classes to the mix. There are also functions
for analysis and making things look nice.
Let’s get rolling: source the vulgaris dataset, which contains
location information for Syringa Vulgaris (the Lilac) observation
stations and US states. This code plots the states and vulgaris
points.
data("vulgaris") #load data par = (mar = c(2,0,0,0)) #set margins of plot area plot(us_states) plot(vulgaris, add = T, pch = 20)
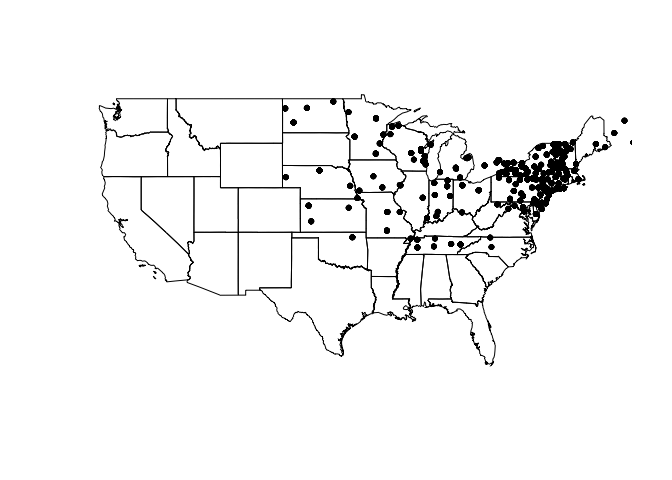
One thing to note here is the structure of these objects. us_states is
a SpatialPolygonsDataFrame, which stores information for plotting shapes
(like a shapefile) within its attributes. vulgaris by contrast is a
SpatialPointsDataFrame, which contains data for plotting individual
points. Much like a data.frame and $, these objects harbor
information that can be accessed via @.
kable(head(vulgaris@data))
| Station | Year | Type | Leaf | Bloom | Station.Name | State.Prov | Lat | Long | Elev | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3695 | 61689 | 1965 | Vulgaris | 114 | 136 | COVENTRY | CT | 41.8 | -72.35 | 146 |
| 3696 | 61689 | 1966 | Vulgaris | 122 | 146 | COVENTRY | CT | 41.8 | -72.35 | 146 |
| 3697 | 61689 | 1967 | Vulgaris | 104 | 156 | COVENTRY | CT | 41.8 | -72.35 | 146 |
| 3698 | 61689 | 1968 | Vulgaris | 97 | 134 | COVENTRY | CT | 41.8 | -72.35 | 146 |
| 3699 | 61689 | 1969 | Vulgaris | 114 | 138 | COVENTRY | CT | 41.8 | -72.35 | 146 |
| 3700 | 61689 | 1970 | Vulgaris | 111 | 135 | COVENTRY | CT | 41.8 | -72.35 | 146 |
Let’s take a look at some functions that use this data.
newVulgaris kable(head(data.frame(newVulgaris)))
| x | y | |
|---|---|---|
| 3 4896 | -67.65 | 44.65 |
| 3 4897 | -67.65 | 44.65 |
| 3 4898 | -67.65 | 44.65 |
| 3 4899 | -67.65 | 44.65 |
| 3 4900 | -67.65 | 44.65 |
| 3 4901 | -67.65 | 44.65 |
gIntersection, as you may have guessed from the name, returns the
intersection of two spatial objects. In this case, we are given the
points from vulgaris that are within us_states. However, the rest of
the vulgaris data has been stripped from the resulting object. We’ve
got to jump through a couple of hoops to get that information back.
<br />newVulgaris <- data.frame(newVulgaris) tmp <- rownames(newVulgaris) tmp <- strsplit(tmp, " ") tmp <- (sapply(tmp, "[[", 2)) tmp <- as.numeric(tmp) vdf <- data.frame(vulgaris) newVulgaris <- subset(vdf, row.names(vdf) %in% tmp)
| Station | Year | Type | Leaf | Bloom | Station.Name | State.Prov | Lat | Long | Elev | Long.1 | Lat.1 | optional | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3695 | 61689 | 1965 | Vulgaris | 114 | 136 | COVENTRY | CT | 41.8 | -72.35 | 146 | -72.35 | 41.8 | TRUE |
| 3696 | 61689 | 1966 | Vulgaris | 122 | 146 | COVENTRY | CT | 41.8 | -72.35 | 146 | -72.35 | 41.8 | TRUE |
| 3697 | 61689 | 1967 | Vulgaris | 104 | 156 | COVENTRY | CT | 41.8 | -72.35 | 146 | -72.35 | 41.8 | TRUE |
| 3698 | 61689 | 1968 | Vulgaris | 97 | 134 | COVENTRY | CT | 41.8 | -72.35 | 146 | -72.35 | 41.8 | TRUE |
| 3699 | 61689 | 1969 | Vulgaris | 114 | 138 | COVENTRY | CT | 41.8 | -72.35 | 146 | -72.35 | 41.8 | TRUE |
| 3700 | 61689 | 1970 | Vulgaris | 111 | 135 | COVENTRY | CT | 41.8 | -72.35 | 146 | -72.35 | 41.8 | TRUE |
Look familiar? Now we’ve got a data frame with the clipped vulgaris
values and original data preserved.
vulgarisSpatial ``` After storing our clipped data frame as a SpatialPointsDataFrame, we can again make use of it - in this case we add a shading scheme to the `vulgaris` points. ``` r shades shades$cols plot(us_states) choropleth(vulgarisSpatial, vulgarisSpatial$Elev,shading = shades, add = T, pch = 20)
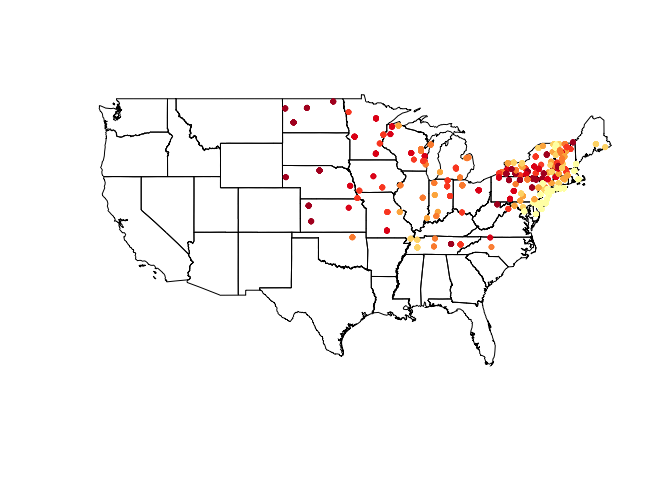
Colors are pretty, but what do they mean? Let’s add a legend.
us_states@bbox #Get us_states bounding box coordinates.
##min max ## r1 -124.73142 -66.96985 ## r2 24.95597 49.37173
plot(us_states) choropleth(vulgarisSpatial, vulgarisSpatial$Elev,shading = shades, add = T, pch = 20) par(xpd=TRUE) #Allow plotting outside of plot area. choro.legend(-124, 30, shades, cex = .75, title = "Elevation in Meters") # Plot legend in bottom left. Takes standard legend() params.
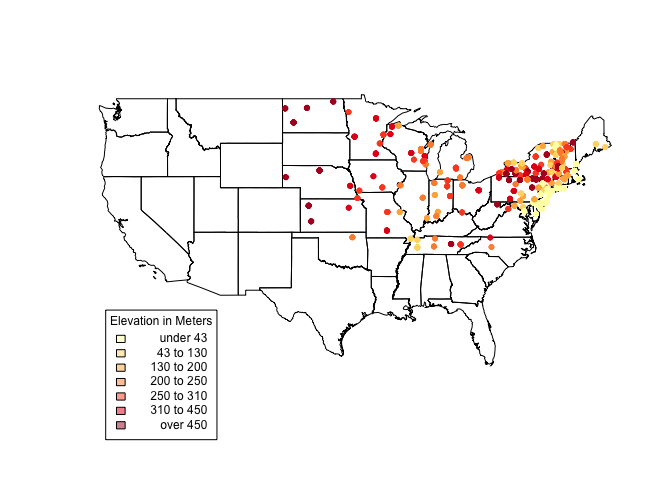
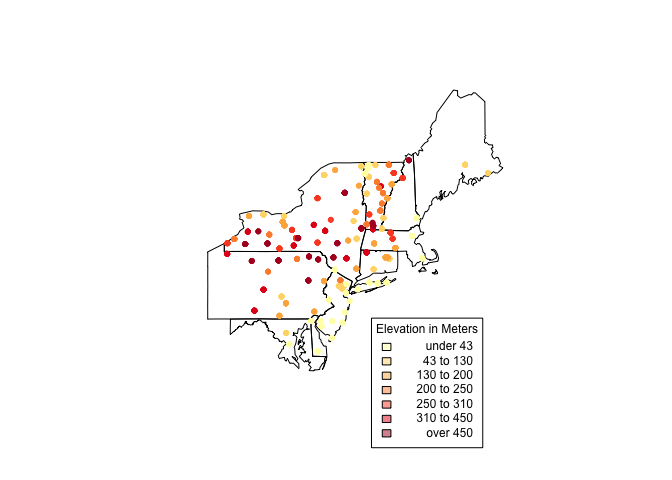
It looks like there’s a lot going on in the Northeastern states. For a
closer look, create another clipping (like above) and plot it. Using the
structure below, we can create a selection vector. I have hidden the
full code since it is repetitive (check GitHub for the full code.)
index '...'
plot(us_states[index,]) choropleth(vulgarisNE, vulgarisNE$Elev,shading = shades, add = T, pch = 20) par(xpd = T) choro.legend(-73, 39.75, shades, cex = .75, title = "Elevation in Meters")
Hopefully this has been a useful introduction (or refresher) on spatial
data. I always learn a lot in the process of writing these posts. If you
have any ideas or suggestions please leave a comment or feel free to
contact me!
Happy mapping,
Kiefer
Vielen lieben Dank für den Einstieg. Leider wird die GIS-Funktion in R von sehr vielen mächtig unterschätzt. Ein schöner Einstieg 🙂
LikeLike
An excellent article. I am new to R and GIS. I always wanted to learn R and GIS for sure as its importance and applications in wildlife ecology and conservation are enormous. I entertain the idea of mapping the distribution of wild Bengal tiger (Panthera tigris tigris) population in Sundarbans mangrove ecosystem in Bangladesh. However, I simply do not know how I can incorporate Sundarbans map in R. Broadly speaking, my question is, if I like to incorporate a map of any ecosystems in R, how I go about it? Surely it is not as simple as scanning a map of Sundarbans in jpeg format and incorporate it in R to eliminate all the unwanted parts of the map and reproduce it as simple line map like you have shown in this article. Would appreciate your kind thoughts in this regard.
Saludos!
Ashraf
LikeLike
Hey Ashraf,
It would be helpful to know what data you have and what format it is in. People definitely use R and spatial analysis to learn more about the movements of animals.
Feel free to send me an email!
Kiefer Smith
LikeLike
Also, here is a link to some info you may find useful: https://cran.r-project.org/web/packages/ctmm/vignettes/variogram.html
LikeLike
Greetings Smith:
Thanks for the reply and good set of questions indeed. As you may know considerable studies have been done on endangered vertebrates especially wild cats like Tigers in Bangladesh or Mountain Lion in United States. Hence lot of data are available from peer-reviewed journal articles and high profile NGOs like World Wildlife Fund and Wildlife Conservation Society. Nevertheless, based on the data set that are readily available, what I intend to do is generating a GIS map focusing three aspects: 1. Distribution based on identification of three distinct mangrove species in the Sundarbans ecosystem 2. Tiger distribution based on direct and indirect counts of tiger numbers 3. Tigers prey (one particular species of deer called Chital deer or Axis axis) distribution based on direct count. Of course data set in these three aspects are scattered across journal and Ph.D papers that I have at my disposal hence surely I can pre-process these data in .CSV format. My limitation is how I can actually generate a map that can neatly layer (superimpose may be another word) these three aspects hence can produce a GIS map that can be updated as we have more data in these three aspects in future. Hence the GIS map can essentially reflect the changes of mangrove tree covers, tiger distribution and deer distribution dynamically over time that is both in spatial and temporal scale.
Of course I am also interested to carry out regression analysis based on these multivariate data set (mangrove trees, tiger numbers and deer numbers). And my ultimate goal is two-pronged. 1. Neat GIS map of Sundarbans mangrove ecosystem 2. GIS map is dynamic hence reflecting both spatial and temporal changes of mangroves, tigers and deer for the past 10 years as an example based on the data set that is linked with the GIS map hence every time I am changing the numbers in my data set, the GIS map gets automatically updated.
Of course all these above are my wishful thinking and at this point whether these can be actually done in R and if so, whether I can gain necessary skills (possibly with your kind collaboration to jointly carry out this Sundarbans tiger project with the aim of scientific publication) are two different matter.
It is pretty evident that you have done your homework and gained considerable skills in R programming language and statistics. This site is testimony to that indeed. Hence it would be great please to do collaborative works on wildlife ecology and conservation. The site I run is called Species Ecology and hotspot link is http://www.speciesecology.wordpress.com. Please take a look at the site and our members. Species Ecology can only be benefited by having someone like you our North American Chapter member for sure. It would give us the opportunity for further collaboration and share your valuable works with our readers for sure. Let me know your thought on that. Also, if you wish to know more about myself and my work, please visit my site too which is http://www.equatorialbiomes.wordpress.com
Apology for lengthy post and thanks for your kind attention.
Saludos
Ashraf
LikeLike
If you have coordinates (or other location data) for each of the variables you are trying to track (tigers, mangroves, prey) it would absolutely be possible to use that information to seek relationships. R has a number of great mapping packages (leaflet comes to mind) that you could use to dynamically plot the data. I might be interested in assisting you with the programming! Would be very cool to be a part of your publication.
Thanks,
Kiefer Smith
LikeLike